Advances in machine learning (ML) have shown great promise for assisting in the work of healthcare professionals, such as aiding the detection of diabetic eye disease and metastatic breast cancer. Though high-performing algorithms are necessary to gain the trust and adoption of clinicians, they are not always sufficient—what information is presented to doctors and how doctors interact with that information can be crucial determinants in the utility that ML technology ultimately has for users.
The medical specialty of anatomic pathology, which is the gold standard for the diagnosis of cancer and many other diseases through microscopic analysis of tissue samples, can greatly benefit from applications of ML. Though diagnosis through pathology is traditionally done on physical microscopes, there has been a growing adoption of “digital pathology,” where high-resolution images of pathology samples can be examined on a computer. With this movement comes the potential to much more easily look up information, as is needed when pathologists tackle the diagnosis of difficult cases or rare diseases, when “general” pathologists approach specialist cases, and when trainee pathologists are learning. In these situations, a common question arises, “What is this feature that I’m seeing?” The traditional solution is for doctors to ask colleagues, or to laboriously browse reference textbooks or online resources, hoping to find an image with similar visual characteristics. The general computer vision solution to problems like this is termed content-based image retrieval (CBIR), one example of which is the “reverse image search” feature in Google Images, in which users can search for similar images by using another image as input.
Today, we are excited to share two research papers describing further progress in human-computer interaction research for similar image search in medicine. In “Similar Image Search for Histopathology: SMILY” published in Nature Partner Journal (npj) Digital Medicine, we report on our ML-based tool for reverse image search for pathology. In our second paper, “Human-Centered Tools for Coping with Imperfect Algorithms During Medical Decision-Making” (preprint available here), which received an honorable mention at the 2019 ACM CHI Conference on Human Factors in Computing Systems, we explored different modes of refinement for image-based search, and evaluated their effects on doctor interaction with SMILY.
SMILY Design
The first step in developing SMILY was to apply a deep learning model, trained using 5 billion natural, non-pathology images (e.g., dogs, trees, man-made objects, etc.), to compress images into a “summary” numerical vector, called an embedding. The network learned during the training process to distinguish similar images from dissimilar ones by computing and comparing their embeddings. This model is then used to create a database of image patches and their associated embeddings using a corpus of de-identified slides from The Cancer Genome Atlas. When a query image patch is selected in the SMILY tool, the query patch’s embedding is similarly computed and compared with the database to retrieve the image patches with the most similar embeddings.
 |
| Schematic of the steps in building the SMILY database and the process by which input image patches are used to perform the similar image search. |
| Second example of using SMILY, this time searching for a lobular carcinoma, a specific subtype of breast cancer. |
However, a problem emerged when we observed how pathologists interacted with SMILY. Specifically, users were trying to answer the nebulous question of “What looks similar to this image?” so that they could learn from past cases containing similar images. Yet, there was no way for the tool to understand the intent of the search: Was the user trying to find images that have a similar histologic feature, glandular morphology, overall architecture, or something else? In other words, users needed the ability to guide and refine the search results on a case-by-case basis in order to actually find what they were looking for. Furthermore, we observed that this need for iterative search refinement was rooted in how doctors often perform “iterative diagnosis”—by generating hypotheses, collecting data to test these hypotheses, exploring alternative hypotheses, and revisiting or retesting previous hypotheses in an iterative fashion. It became clear that, for SMILY to meet real user needs, it would need to support a different approach to user interaction.
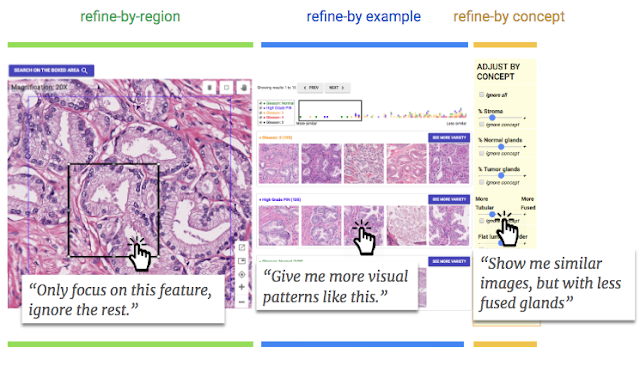
Through careful human-centered research described in our second paper, we designed and augmented SMILY with a suite of interactive refinement tools that enable end-users to express what similarity means on-the-fly: 1) refine-by-region allows pathologists to crop a region of interest within the image, limiting the search to just that region; 2) refine-by-example gives users the ability to pick a subset of the search results and retrieve more results like those; and 3) refine-by-concept sliders can be used to specify that more or less of a clinical concept be present in the search results (e.g., fused glands). Rather than requiring that these concepts be built into the machine learning model, we instead developed a method that enables end-users to create new concepts post-hoc, customizing the search algorithm towards concepts they find important for each specific use case. This enables new explorations via post-hoc tools after a machine learning model has already been trained, without needing to re-train the original model for each concept or application of interest.
Through our user study with pathologists, we found that the tool-based SMILY not only increased the clinical usefulness of search results, but also significantly increased users’ trust and likelihood of adoption, compared to a conventional version of SMILY without these tools. Interestingly, these refinement tools appeared to have supported pathologists’ decision-making process in ways beyond simply performing better on similarity searches. For example, pathologists used the observed changes to their results from iterative searches as a means of progressively tracking the likelihood of a hypothesis. When search results were surprising, many re-purposed the tools to test and understand the underlying algorithm, for example, by cropping out regions they thought were interfering with the search or by adjusting the concept sliders to increase the presence of concepts they suspected were being ignored. Beyond being passive recipients of ML results, doctors were empowered with the agency to actively test hypotheses and apply their expert domain knowledge, while simultaneously leveraging the benefits of automation.
With these interactive tools enabling users to tailor each search experience to their desired intent, we are excited for SMILY’s potential to assist with searching large databases of digitized pathology images. One potential application of this technology is to index textbooks of pathology images with descriptive captions, and enable medical students or pathologists in training to search these textbooks using visual search, speeding up the educational process. Another application is for cancer researchers interested in studying the correlation of tumor morphologies with patient outcomes, to accelerate the search for similar cases. Finally, pathologists may be able to leverage tools like SMILY to locate all occurrences of a feature (e.g. signs of active cell division, or mitosis) in the same patient’s tissue sample to better understand the severity of the disease to inform cancer therapy decisions. Importantly, our findings add to the body of evidence that sophisticated machine learning algorithms need to be paired with human-centered design and interactive tooling in order to be most useful.
Acknowledgements
This work would not have been possible without Jason D. Hipp, Yun Liu, Emily Reif, Daniel Smilkov, Michael Terry, Craig H. Mermel, Martin C. Stumpe and members of Google Health and PAIR. Preprints of the two papers are available here and here.


No comments:
Post a Comment